mRNA-Based Gene Editing Therapeutics
Genome editing has been gaining importance since the last decade as a potential alternative to gene therapy. Nucleases (ZFNs, TALENs, etc.) and the CRISPR–Cas9 system provide powerful tools for site-specific modification of genomes. However, these approaches have the risk of nonspecific editing as the DNA-based vectors translate the enzymes involved. In-vitro transcribed mRNA (IVT mRNA) encoding these enzymes can be applied to edit or modify the genome to address this issue. The transient expression of the gene-modifying enzymes from the mRNA would reduce the off-targeting effects, as the enzymes are required only for a short duration of time during the process. The mRNA-based gene editing comes with low mutation risk and would provide a controlled and precise therapeutic effect.
Technology Overview
Messenger RNA drugs will modify the genetic blueprint of a cell using artificial endonucleases, such as ZFNs, TALENs, and CRISPR/Cas nuclease systems. Among these, CRISPR/Cas9 systems are most frequently being used for gene editing due to the ease of design and implementation. T cells are being transfected with mRNA-encoding endonucleases through electroporation for therapeutic applications such as treating cancer and infectious diseases. CRISPR/Cas9 mRNA-disrupted allogeneic CAR T cells showed efficient antitumor activity in vitro and in vivo. These mRNA-based gene editing techniques have also been explored for the generation of stem cells for many therapeutic and clinical applications. Recently, HIV-1 Tat mRNA was delivered into bone marrow mesenchymal stem cells (BMSCs), confirming the inhibitory effect of HIV-1 Tat protein on the hematopoietic support function of hBMSCs. A lot of efforts have also been made for gene editing of induced pluripotent stem (iPS) cells, as it holds great significance in cell therapy and disease modeling.
Future efforts will pay attention to safe and efficacious delivery strategies of mRNA for further therapeutic purposes.

Figure 1. mRNA Drugs for Various Disease Indications
Approaches for mRNA Delivery
The key factor determining the success of genome editing is the effective delivery of the mRNA encoding the gene-editing enzymes.
There are various barriers associated with the delivery, which include cytotoxicity caused by manipulation and delivery vectors, cellular uptake, endosomal escape, mRNA degradation triggered by lysosome and cytoplasmic nucleases, immune response mediated through the pattern-recognition receptors of the innate immune system (such as Toll-like receptors and retinoic acid-inducible gene I (RIG-I)-like receptors), reduced manufacture capacity of delivery formulations and poor output rate of the delivery process.
mRNA can be delivered to the cells using physical methods and non-viral synthetic materials. Physical methods include electroporation, microinjection, and mechanical deformation (such as squeeze).
A large number of nanomaterials are not being developed for mRNA delivery in cells. Lipid nanomaterials are being extensively explored for transferring RNA molecules. Cationic polymers such as polyethylenimine (PEI) are also widely used for in-vivo transfection of mRNA.
Applications of IVT mRNA for Gene Editing
For Research:
IVT mRNA encoding gene editing enzymes find its application widely in research and preclinical trials. It is applied in cell engineering to modify cells such as human T cells, HPSCs, and other cells through electroporation of Cas9 mRNA, Zinc finger nucleases, and TALEN. Microinjection of the mRNA encoding these enzymes into one-cell-stage embryos or zygotes has also been explored for developing various animal models, including zebrafish, mice, and pigs. For instance, to advance genome-editing therapies for AIDS, researchers can track edited primary human hematopoietic stem and progenitor cells (HSPCs) in a mutation-specific manner by transfecting macaque-specific CCR5 ZFN mRNA ex vivo into them and then engrafting the modified HSPCs in a large animal model. Genetically modified rodents are being generated utilizing the CRISPR-Cas9 system by microinjection of Cas9 mRNA and sgRNA in cells in various universities and companies.
In Clinical Trials:
Several clinical trials using mRNA-based gene editing systems are in progress.
- Sangamo Therapeutics developed a study for HIV treatment by using SB-728mR, mRNA of specialized ZFNs targeting the CCR5 gene in HSPCs
- In another study, SB-728mR was used to modify the T cells
- Intellia Therapeutics has initiated its Phase 3 trial of NTLA-2001, a single-dose CRISPR-based treatment to inactivate the TTR gene for treating transthyretin amyloidosis with cardiomyopathy
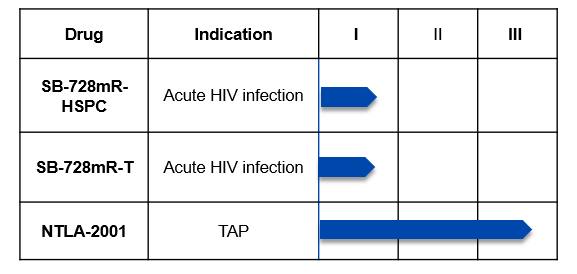
Figure 2. Assets in Clinical Trials
Key Players and Their Recent Activities
Moderna and Life Edit Therapeutics have partnered to explore and develop in vivo mRNA gene editing therapies. Life Edit’s proprietary gene editing technologies, along with Moderna’s mRNA platform, will be used to discover the therapies against a select set of therapeutic targets.
Intellia Therapeutics and ReCode Therapeutics have recently collaborated to develop novel gene editing therapies for cystic fibrosis, wherein they will be utilizing Intellia’s proprietary CRISPR-based gene editing platform and ReCode’s proprietary Selective Organ Targeting (SORT) lipid nanoparticle (LNP) delivery platform to correct CF disease-causing gene mutations.
Sangamo BioSciences has lipid nanoparticle technology for delivering mRNA of Sangamo’s proprietary zinc finger nucleases (ZFNs) to explore numerous new therapeutic opportunities.

Figure 3. Key Players in mRNA-based Gene Editing Technology
Conclusion
Gene editing utilizes programmable nucleases for insertion/deletion mutations of the genes to treat diseases such as cancer, infectious diseases, hematological diseases, and immune system disorders. Messenger RNA drugs are an emerging class of drug therapy that has the potential to play a role in gene therapy, wherein mRNA can be used to deliver these programmable nucleases using in vitro–transcribed mRNA (IVT mRNA) molecules. Effective transfection of the mRNA for gene editing has been achieved for the three most essential nucleases, namely the clustered regularly interspaced short palindromic repeat (CRISPR)-associated protein (CRISPR/Cas) nuclease system, zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs). Keeping in mind the amount of research and clinical trials going on in this field, it is evident that IVT mRNA-based holds enormous potential in therapeutic applications in the future.